Ewen Callaway wrote an excellent review of the state of our knowledge concerning the D614G variant of the SARS-CoV-2 Spike protein: https://www.nature.com/articles/d41586-020-02544-6
You are browsing archives for
Ewen Callaway wrote an excellent review of the state of our knowledge concerning the D614G variant of the SARS-CoV-2 Spike protein: https://www.nature.com/articles/d41586-020-02544-6

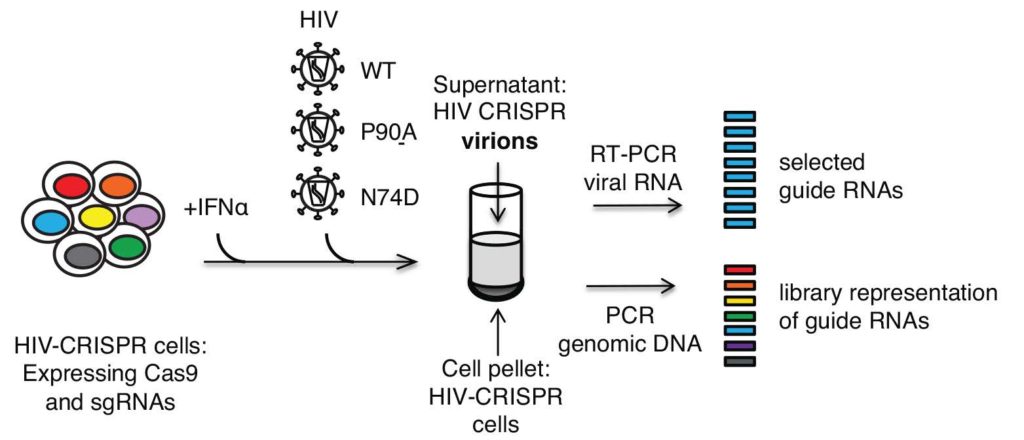
Our manuscript, “TRIM34 acts with TRIM5 to restrict HIV-1 and SIV capsids”, is now online at bioRxiv. This manuscript is the result of a great tri-lab collaboration.
The story springs from a CRISPR loss-of-function screen that was brilliantly designed and expertly conducted by Molly Ohainle in the lab of lab of Michael Emerman at the Hutch in Seattle. Kyusik Kim from my lab at UMass Medical School confirmed the role of these TRIM proteins in primary human blood cells. Our wonderful collaborator Sevnur Komurlu in Ed Campbell’s lab at Loyola Univ in Chicago provides the groovy images.
Kyusik Kim, Ann Dauphin, Sevnur Komurlu, Sean M. McCauley, Leonid A. Yurkovetskiy, Claudia Carbone, William E. Diehl, Caterina Strambio-De-Castillia, Edward M. Campbell, and Jeremy Luban. Cyclophilin A protects HIV-1 from restriction by human TRIM5α. Nature Microbiology is online today and the full PDF can be accessed here.
In 1993, in one of the first two-hybrid screens of a mammalian cDNA library for encoded proteins that interact with a protein of interest, undergraduate student Karen Bossolt discovered that the HIV-1 Gag polyprotein, as well as the mature capsid protein (CA) embedded within it, bind to the cellular protein cyclophilin A (CypA). In 2001, graduate student Douglas Braaten formally demonstrated the importance of this interaction for HIV-1 replication in what was perhaps the first targeted gene disruption experiment by homologous recombination in somatic cells. Over the years, attempts to determine the mechanism of action of cyclophilin A have nonetheless taken many unexpected turns.
Undergraduate student Ettaly Franke found that Cyclophilin A was incorporated into HIV-1 virions at a molar ratio ~1:5 with respect to CA. This suggested that cyclophilin A played a role during virion assembly or within the virion itself. This idea was supported by the observation that infectivity decreased when virions were produced in the presence of cyclosporine, a competitive inhibitor of the Gag-CypA or CA-CypA interaction. Ultimately, graduate student Elena Sokolskaja demonstrated that the effects of cyclosporine during virion assembly were independent of the CA-CypA interaction and could be attributed to disruption of Env glycoprotein incorporation into particles. Elena also showed that HIV-1 CA interaction with TARGET CELL cyclophilin A was important for HIV-1 infectivity. Still today, it remains an open question whether cyclophilin A plays any role during HIV-1 virion assembly, or as a virion component.
Several observations suggested that cyclophilin A protected the incoming HIV-1 CA core from a dominant-acting, antiviral factor within human target cells. Among other properties, the putative antiviral factor was cell type-specific, species-specific, saturated by large amounts of susceptible virion cores, and disrupted by arsenic. Then, in 2004, two screens identified TRIM5 as a species-specific, capsid-specific, restriction factor. The first screen, which used a macaque cDNA expression library, was conducted by graduate student Matt Stremlau in the Sodroski lab. He showed that the macaque TRIM5α orthologue restricted HIV-1; he was unable to detect this anti-HIV-1 activity with the human TRIM5α orthologue. The second screen was conducted by graduate student David Sayah who screened an owl monkey cDNA library. David discovered that the potent anti-HIV-1 restriction activity in this species was also due to the TRIM5 gene, but with a twist: a CypA cDNA had retrotransposed into the owl monkey TRIM5 locus to create a TRIM5-CypA fusion gene that is among the most potent anti-HIV-1 restriction factors. Amazingly, there is now evidence that CypA cDNA has retrotransposed into the TRIM5 locus at least 9 separate times over the course of phylogeny to create fusion genes. Presumably these TRIM5-CypA fusion genes were selected for by challenges with lentiviral ancestors of HIV-1.
The above experiments suggested that, in human cells, CypA might protect HIV-1 from restriction by TRIM5α. Experiments conducted by Elena in the early days of RNAi indicated that the effects of CypA and TRIM5α were independent of each other when HIV-1 infection was measured on tumor cell lines. Since then, working on a range of different projects, a line of graduate students that includes Thomas Pertel, Nadia Rahm, Martha Neagu, Alberto De Iaco, and Sean McCauley, have improved the efficiency of lentiviral vector-based reverse genetic tools for gene disruption, and for gene delivery, in primary human blood cells. Most recently, graduate students Kyusik Kim and Sean McCauley developed what are the most efficient lentiviral vectors yet.
While studying unrelated host factors that he hypothesized to regulate HIV-1 infection, Kyusik Kim fortuitously disrupted CypA in primary human dendritic cells; CypA had been intended as a negative control since he expected it to have little effect in that specific experiment. To his surprise, CypA knockdown had a qualitatively bigger effect on HIV-1 replication in primary human dendritic cells than it had had in the tumor cell lines that had been used previously. Prior to this, our lentiviral vectors were not efficient enough to disrupt CypA to this extent in dendritic cells. Kyusik found the same impressive CypA knockdown phenotype in primary human macrophages and CD4+ T cells. As described in his new paper, Kyusik shows pretty conclusively that HIV-1 replication is very dependent on CypA in primary cells. But then, most surprising, and perhaps most important of all, Kyusik found that CypA protects HIV-1 from the potent restriction activity of endogenous human TRIM5α in these cells. Until now, the human orthologue of TRIM5α was thought to have little effect on HIV-1 replication.
Congratulations to Kyusik and his collaborators, Ann, Sevnur, Sean, Leonid, Claudia, Ted, Caterina, and Ed!!!!

Yu, T., Koppetsch, B.S., Pagliarani, S., Johnston, S., Silverstein, N.J., Luban, J., Chappell, K., Weng, Z., and Theurkauf, W.E. (2019). The piRNA Response to Retroviral Invasion of the Koala Genome. Cell. is online today!!!
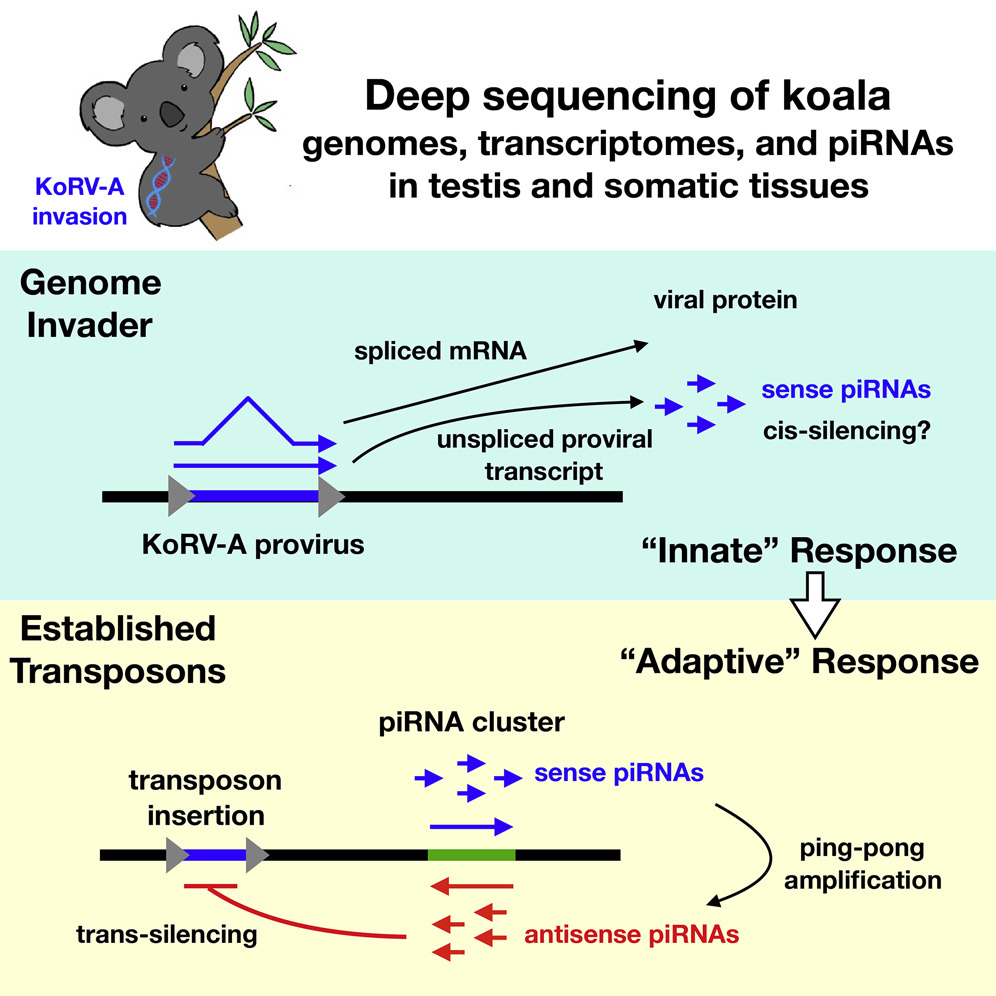
Fruit fly researcher Bill Theurkauf, our neighbor here in the Program in Molecular Medicine at UMass Medical School, along with UMass Med bioinformatics guru Zhiping Weng, has been studying the mechanisms by which piRNAs are generated in the germ cells of Drosophila. It is believed that the piRNAs are a sequence-specific defense system that protects the germ cell genome from DNA-damage caused by jumping genes like endogenous retroviruses. Similar piRNAs are found in mice and humans, and it is important to remember that 8% of the human genome is composed of retrovirus sequence.
Bill heard that there was an epidemic in Koalas caused by KoRV-A, a retrovirus that causes an immunosuppressive illness with parallels to AIDS. What caught Bill’s interest was the fact that, unlike HIV-1, KoRV-A was not only transmitted horizontally, but also vertically through the germline.
As described in a manuscript that was published today, Bill then teamed up with colleagues in Brisbane to determine whether the germ cells of infected Koalas generated piRNAs specific for KoRV-A. Indeed they do, though, unlike “mature” piRNAs, the piRNAs targeting KoRV-A were only + sense-stranded. This suggests that the piRNA system was acting as an antiviral system that acts directly on the KoRV-A transcripts to degrade them.
Another interesting detail is that only unspliced KoRV-A transcripts were detected among the piRNAs, despite 5-fold higher levels of the spliced transcripts. This suggests that the unspliced transcript, a molecular feature typical of all retro-elements is a PAMP, or danger signal, that is recognized by our cells antiviral machinery. Similar ideas about unspliced transcripts from retroelements have been reported by the lab of Hiten Madhani regarding siRNAs in Cryptococcus, and, separately, by the Gummuluru Lab and by the Luban Lab concerning unspliced transcripts of HIV-1 in human immune cell types.
Noah Silverstein, graduate student in the Luban Lab, contributed to the piRNA manuscript by identifying and collecting mouse strains bearing endogenous retroviruses in their genome that are close cousins to KoRV-A. Like the koalas, these mice also generated +-stranded piRNAs in their germ cells.
The Theurkauf, Weng, and Luban Labs here at UMass Medical School, along with our Brisbane colleagues Johnston and Chappell, will continue working to address the many questions that remain to be answered about these piRNAs targeting retroviruses that are newly integrated into the mammalian genome.
Our paper is covered in an article in the New York Times, on the website for UMass Medical School, and on local TV in Brisbane.
Leonid Yurkovetskiy has been selected as a 2019 Award Recipient of the Charles A. King Trust Postdoctoral Fellowship Program.
Given the potential of his research project to further understanding of the fundamental biology of cancer, he was specifically awarded a 2019-2021 Sara Elizabeth O’Brien Trust Fellowship within the King Program.
The title of Lonya’s project is, “Mechanism of epigenetic silencing by the HUSH complex.”
The project springs from his discovery that the primate immunodeficiency virus accessory proteins Vpr and Vpx bind to and promote the DCAF1-Cul4A-ubiquitin-dependent degradation of the HUSH transcriptional silencing complex. The background for the experiments proposed in his fellowship can be found in his Nature Microbiology manuscript.
We’re much obliged to the reviewers of the fellowship applications, to The Charles A King Trust, and to the Sara Elizabeth O’Brien Trust.

Our neighbor here in the Program in Molecular Medicine, at the University of Massachusetts Medical School, Paul Greer, has received an NIH Director’s New Innovator Award. Given that he just started his own lab, and given how innovative we know him to be, we think this New Innovator Award is well deserved. Congrats to Paul!!!

Our manuscript entitled, “Influence of different glycoproteins and of the virion core on SERINC5 antiviral activity”, is now online as a preprint at bioRxiv.
The authors were : William E. Diehl, Mehmet H. Guney, Pyae Phyo Kyawe, Judith M. White, Massimo Pizzato, Jeremy Luban
doi: https://doi.org/10.1101/780577
In 2015, we reported that HIV-1 Nef blocks the antiviral host factor SERINC5, and that the structurally unrelated MLV Glycogag acts similarly to Nef (Nature 526:212). The HIV-1 glycoprotein was identified as a viral determinant of susceptibility to restriction. In contrasts, GPs that fuse in acidic endosomes, i.e., VSV and EBOV, were resistant to restriction. Virion membrane-associated SERINC5 blocks an early step in the infection cycle but the exact mechanism of restriction has not been elucidated.
The new manuscript examined a large panel of GP pseudotypes on three types of retrovirion cores and makes the following observations:
1. we identify the first non-retroviral GPs that are sensitive to SERINC5 restriction
2. GP requirement for acidification does not necessarily make the particle SERINC5-resistant.
3. the core of the virion influences sensitivity to restriction.
Our findings extend understanding of the requirements for SERINC5 restriction, potentially expand the significance of SERINC5 beyond retroviruses, and provide useful information to those attempting to understand the SERINC5 restriction mechanism.
@NoahSilverstein, the #VibrantVermontVocalist, is now an #NIHBankrolledWorcesterScientist
Grant Number: 1 F30 HD100110-01A1
Principal Investigator: Silverstein, Noah Jacob
Project Title: Mechanism of epigenetic inheritance in a mouse model of acute paternal stress
Institution: UNIV OF MASSACHUSETTS MED SCHOOL
@UMassMedical @ProgMolMed
#EpigeneticInheritance
#ResistanceToInfluenza
#Bravissimo & Congrats

The manuscript resulting from our collaborative Ebola virus project with the Sabeti, Andersen, and Rambaut Labs, has been published, back-to-back with a highly complementary paper from a group led by Jonathan Ball, along with a brilliant commentary from Trevor Bedford and Harmit Malik. Since our manuscript went on line it’s had a bit of press coverage:
The New York Times
Science Magazine
The Atlantic
NPR – All Things Considered
BBC Science in Action (audio mp3, starts at 7’45” after the Italian earthquake story)
BBC News
The Washington Post
Boston Herald
Scientific American
Scientific American Podcast
Time
This Week in Virology (TWiV) Podcast
NBC news
New Scientist
Worcester Telegram and Gazette
UMassMed Now (video)
UMassMed News
Channel 3 New Worcester (YouTube news report video, starts at 6’09”)
CIDRAP News and Perspective. Ctr for Infect Dis Res and Policy, Univ of Minnesota
The Huffington Post
AllAfrica
Newsela
ZME Science
The Hindu
The Scripps Research Institute Online Weekly
Counsel and Heal
Express.co.uk
Oswego News
UPI
International Business Times
Bismark Tribune
Galileo Net
Science Daily
IFL Science
Science Blog
Sputnik News
Le Scienze
Romper
Wired.co.uk
Homeland Security News Wire
Answers Africa
Crofsblogs
Inquisitr
Digital Journal
Microbes
iTech Post
Virology.ws
Tech Times
Contagion Live
Telegiz
Earth-Chronicles
Kaiser Health News
Top247
Health News NG
Ultima Voce
People Magazines
Headlines-News
Student Newspaper
BMC Medicine
SG.News.Yahoo
Garudapro
Andalou Agency
Celebstarznews
Nottingham.ac.uk
Focus Online
Daykan
Ooyuz
Vounbeyeu
Antizdrav
ProMED-mail
Medline Plus
Wired.it