HIV-1 nef and the SERINC family of restriction factors
A podcast conversation concerning the following text was recorded for This Week in Virology (TWiV episode #409) on September 30th, 2016. Please click here if you wish to listen to it.
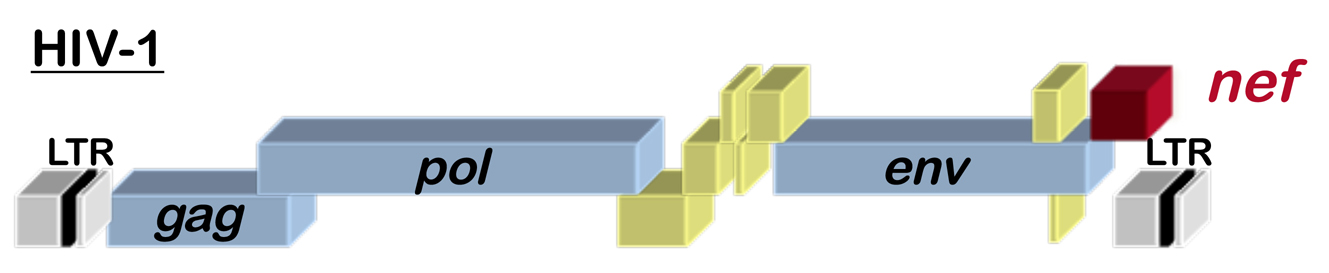
Fig. 1 Schematic of the HIV-1 provirus
HIV-1, the virus that causes AIDS, has 9 genes, including the accessory gene called nef (see the location of nef within the HIV-1 genome in Figure 1). HIV-1 Nef is perhaps one of the best studied proteins in history, with thousands of papers in PubMed. Among the many functions that have been reported for it, Nef alters cellular membrane trafficking, and, by so doing, downregulates cell surface proteins such as CD4, the primary HIV-1 receptor. Given the size of the Nef literature, we hope that all the many Nef experts out there will accept our apologies for the limited story that follows.

Fig 2. Ron Desrosiers, Jeff Lifson, and Hung Fan, in typical virologist attire.
Once upon a time, the New England Primate Research Center (NEPRC) in Southborough, Massachusetts made major contributions to our understanding of HIV-1 and AIDS (the NEPRC closed in 2015). For 12 years, a Harvard University Professor named Ronald Desrosiers served as the Director of the NEPRC. Ron is now Professor of Pathology at the University of Miami. Importantly, while at the NEPRC, Ron developed a reputation for wearing outrageous Madras plaid jackets, shirts, pants, and shorts (Figure 2). Among other things, Ron studied SIV, a close cousin of HIV-1 that causes an AIDS-like disease in rhesus macaques. Ron and his colleagues found that, when nef was deleted from cloned SIV, the virus was still able to infect macaques, but the viremia was lower than with the wild type and the disease course was greatly slowed (Kestler et al., 1991).
One day in 1995, in a pub in Worcester, Massachusetts, Ron had a beer or two with his good friend John Sullivan (Figure 3), a prominent AIDS researcher at the University of Massachusetts Medical School, as well as an avid golfer. John told Ron about people who, despite having been infected with HIV-1 for more than a decade, remained AIDS-free, without taking antiviral drugs. After leaving the bar (and perhaps some other hurdles), they had the virus that infected these people sequenced and found that the nef gene in one of them was defective (Kirchhoff et., 1995). With respect to disease, then, the nef-defective HIV-1 was behaving similarly to the nef-defective SIV that Ron had studied at the NEPRC. An impressive cluster of people with non-progressive HIV-1 infection was also identified in Sydney, Australia; a blood donor infected with nef-mutant HIV-1 had donated the virus to at least 6 blood recipients (Deacon et al., 1995).

Fig 3. John Sullivan and Ron Desrosiers
At the same time, back then in 1995, several labs reported that HIV-1 nef acted within the virion producer cell to increase the infectivity of the virions that are produced (Chowers et al., 1995; Aiken and Trono, 1995; Schwartz et al., 1995; Goldsmith et al., 1995). In other words, virions produced in the absence of nef are less infectious (Figure 4). The stage in the viral replication cycle where the replication defect becomes manifest has not been easy to pinpoint (see text that follows). Biochemical characterization of virions produced in the absence of nef failed to identify obvious defects that could be reproduced by multiple labs. It was clear though, that the block to replication occurs very soon after virions generated in the absence of nef bind to a susceptible target cell.
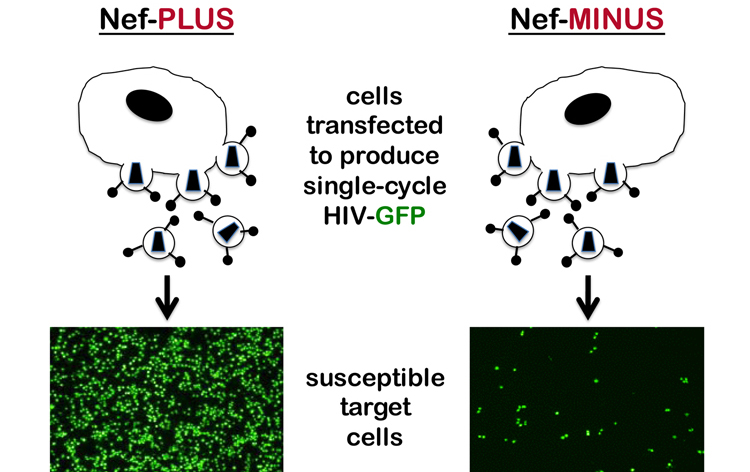
Fig. 4. Nef acts in producer cells to increase virion infectivity
Over the ensuing 20 years a number of important observations made by many people ultimately helped explain how Nef contributes to virion infectivity. Among these people, Massimo Pizzato began working on Nef in 2000, while he was a postdoc in the lab of Heinrich Gottlinger. Notably, Massimo is a highly accomplished clarinetist with a Masters Diploma from the Conservatorio di Musica”Arrigo Pedrollo”, Vicenza, Italia. While with Heinrich, Massimo discovered that Nef binds to Dynamin 2, a GTPase involved in clathrin-mediated endocytosis, and that this interaction is required for the effects of Nef on virion infectivity (Pizzato et al.,2007). Later, as a Lecturer at Imperial College London, and then at the University of Geneva, and now at the University of Trento, Massimo noted that the effect of Nef on virion infectivity was dependent on the cell type that was used to produce the virions. For example, HIV-1 produced from Jurkat T cells in the absence of nef is ~50-fold less infectious than virus produced in the presence of nef, whereas nef has minimal effect when virions are produced from HT1080 cells (see Figure 5 and Rosa et al., 2015).
The cell type-dependence of the Nef phenotype was not particularly surprising in itself, except that Massimo noticed that the magnitude of the Nef phenotype varied enormously, and, most strikingly, when he compared supposedly identical cell lines from different sources. He then realized that as much as 25% of cell lines in wide circulation – even lines distributed by repositories – were contaminated with exogenous retroviruses, either murine leukemia virus or gibbon ape leukemia virus. Amazingly, those cell lines that were contaminated were completely Nef-independent with respect to the production of infectious HIV-1 virions. Massimo then showed that Nef-dependent cells that were free from retrovirus contamination, became Nef-independent if he infected them with Moloney murine leukemia virus (MoMuLV). Deletion analysis showed that the activity within MoMuLV mapped to Glycosylated Gag, a type II transmembrane protein consisting of Gag polyprotein bearing an N-terminal extension, the translation of which starts from an upstream CUG (Pizzato, 2010). The function of Glyco-Gag had been a mystery since Hung Fan (see Figure 2) and others first described it back in the 1970s (Edwards and Fan, 1979). Glyco-Gag is structurally unrelated to Nef but nonetheless substitutes for it functionally. Glyco-Gag and Nef, then, are wonderful examples of convergent evolution.
The effect of Nef on virion infectivity could be explained if Nef substituted for a factor that HIV-1 needed to form an infectious virion; this factor would be lacking in Jurkat but present in HT1080 cells. Alternatively, Nef might act to suppress an antiviral factor that is present in Jurkat but not in HT1080 cells. To determine which was the case, Massimo generated cell fusions (heterokaryons) between Jurkat and HT1080 cells. The heterokaryons were then used to produce virions in the presence or absence of Nef. The phenotype of the virus produced by the heterokaryons was found to be the same as that of the virus produced by Jurkat, indicating that the Jurkat phenotype was dominant (Rosa et al., 2015 and Figure 5). Nef increased HIV-1 infectivity in Jurkat cells by inhibiting an inhibitor of HIV-1 that is present in Jurkat cells.
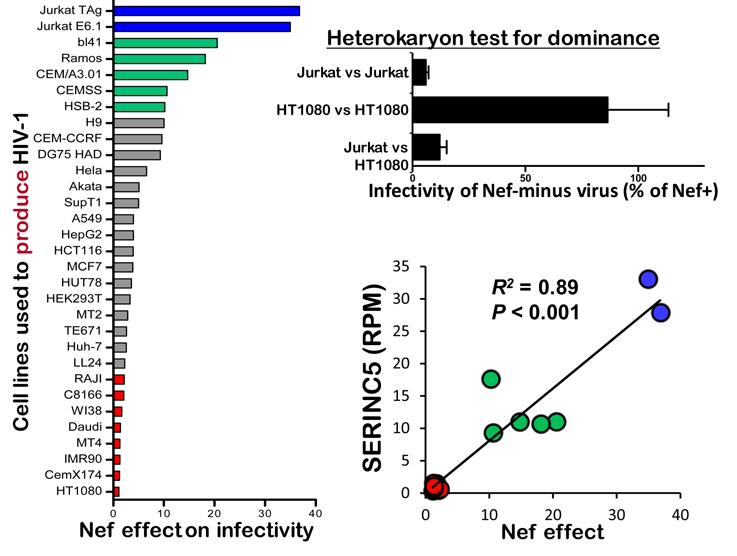
Fig 5. Discovery of SERINC5 as the Nef-inhibited inhibitor of HIV-1 (from Rosa et al., 2015, click on figure for link)
Massimo hypothesized that the level of expression of the putative, nef-sensitive, anti-HIV-1 host factor in a given cell line would correlate with the magnitude of the nef effect on the infectivity of virions produced by that cell. He generated cDNA libraries from the cell lines indicated in blue, green, or red in Figure 5, and, using RNA-Seq, determined the relative expression of all the RNAs in those cells. Federico Santoni then ranked the genes in terms of relative expression (RPM) in a cell line and the magnitude of the nef effect in that cell line. SERINC5 was the highest scoring gene (R2 = 0.89).

Fig 6. Massimo Pizzato and Federico Santoni
At the same time that Rosa et al., 2015 were performing their differential expression screen, unbenownst to them, the group of Heinrich Gottlinger had performed differential mass spectrometry on highly enriched HIV-1 virions produced in the presence or absence of Nef. The only cellular protein that was reproducibly and uniquely associated with virions that were produced in the absence of Nef was SERINC3 (Usami et al., 2015). One fateful day at the 2014 Cold Spring Harbor Retroviruses Meeting, in a sort of perverse game of Molecular Biology Charades, Pizzato and Gottlinger slapped their respective bald heads, mouths agape in astonishment, with the realization that they had each independently discovered the identity of the long-sought, Nef-sensitive, HIV-1 restriction factor. Further experiments by both groups then showed that SERINC5, and to some extent SERINC3, are indeed responsible for the Nef-sensitive anti-HIV-1 activity. Ultimately, efforts to publish the results of the wonderfully complementary screens – one genetic, one biochemical – were coordinated (Rosa et al., 2015; Usami et al., 2015).
Future Questions that we and others are pursuing
1. Which retroviruses are sensitive to inhibition by SERINC5 or other SERINC family members?
2. Other than retroviruses, which enveloped viruses are sensitive to inhibition by SERINCs?
As part of our survey of glycoproteins encoded by different enveloped viruses we tested Ebola virus strains from the West African outbreak of 2013-2016. Though we found no evidence for SERINC5-mediated inhibition of infectivity directed by Ebola virus glycoproteins, Ted Diehl noticed that certain mutants from the epidemic increased the infectivity. These results are described on our website and in a publication.
3. Given the convergent evolution evidenced by Nef and Glyco-Gag, are there other unrelated viral proteins that overcome the antiviral activity of SERINCs?
4. What is the precise mechanism by which SERINC inhibits virion infectivity?
5. Other than the Env glycoprotein, are there other viral determinants of sensitivity to inhibition by SERINC?
6. What is the structure of SERINC5?
7. Are there human polymorphisms in SERINC5 that might be relevant to viral inhibition?
8. What is the normal cellular function of SERINCs?
To be continued…